
Bile acid dependent and independent roles of fibroblast growth factor 15/19 in liver regeneration
The liver has a remarkable capacity to regenerate itself after extensive loss of tissue. This regenerative capacity makes it possible to surgically remove large parts of the liver when a tumor is present (1). Furthermore, liver regeneration also enables specials forms of liver transplantations, for instance living donor liver transplantation (LDLT), where a healthy person donates a portion of his liver to the transplanted patient. However, inadequate liver regeneration can cause serious clinical conditions leading to liver failure and may be fatal for these patients (2). Recovery after partial hepatectomy consists of several proliferative and hepatoprotective signaling cascades. Increased expression of cell cycle progression genes such as Cyclin D1 and Cyclin E1 stimulate quiescent hepatocytes to enter G1 phase. One of the key cytokines that is rapidly induced upon partial hepatectomy is IL-6, which regulates the transcription factor STAT3 that in turn generates a proliferative response in hepatocytes (3).
After losing a large part its total size, the liver has to cope with a bile acid overload already in the first hours after partial hepatectomy. Bile acids are natural ligands of the farnesoid X receptor (FXR) and are closely involved in the process of liver regeneration. Activation of intestinal FXR leads to production and release of fibroblast growth factor 19 (FGF19) (4). Fibroblast growth factors (FGFs) form a large family of structurally related proteins with pleiotropic functions that can act on multiple cell types. For instance, human FGF19, and the murine orthologue FGF15, play an important role in bile acid homeostasis, glucose homeostasis, development and proliferation (5-7). FGF15 and FGF19 are ligands for the fibroblast growth factor receptor 4 (FGFR4) and its co-receptor β-Klotho (KLB), that promotes this binding and is necessary for the functional effects in hepatocytes and other FGFR4 expressing tissues (8). A link between FGF19-FGFR4 and cell proliferation, but also tumorigenesis has been demonstrated in several studies. For instance, FGF15 deficiency impaired liver regeneration in mice (9). Furthermore, both FGFR4 deficiency as well as blocking the interaction of FGF19-FGFR4 interaction decreased hepatocyte proliferation and tumor growth (10-12). Moreover, FGF19 stimulates STAT3 signaling in vivo, which most likely plays a major role of the proliferative effects of FGF19 (13). In addition, FGF15/19 represses bile acid synthesis (14) and bile acid uptake transporters in the liver (15). Using a comprehensive set of transgenic mice models Kong et al. recently determined the role of FGF15 in liver regeneration independent of bile acid levels (Figure 1) (16). They developed a model using the Tet-off system where Fgf15 expression is driven by the FABP1 promoter, resulting in Fgf15 overexpression in liver and intestine. Administration of doxycycline for 5 days resulted in transcriptional repression of the Fgf15 transgene in both intestine and liver. Surprisingly, five days after doxycycline withdrawal, Fgf15 overexpression returned in the intestine but not in liver. This allowed the authors to compare two different models, the vehicle treated mice that overexpress Fgf15 in both hepatocytes and enterocytes, and the doxycycline followed by vehicle treated mice that show intestine-specific overexpression of Fgf15. Worth noting is that Fgf15 in mouse is endogenously detected in enterocytes only, while human FGF19 expression can also be detected in liver during cholestasis or cirrhosis (17). Liver-specific expression of Fgf15 was achieved using viral transduction of Fgf15 knockout mice using adeno-associated viral vectors. This virus has a liver specific tropism when the AAV8 subtype is used. Finally, they directly injected specially modified FGF15 to have systemic exposure without specific tissue-specific FGF15 secretion.
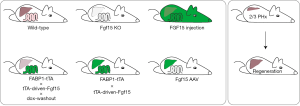
The bile acid pool size is reduced in mice upon increased FGF15 signaling, irrespective of tissue or origin. Notably, this reduction in pool size is not reflected by bile acid levels in plasma, which are increased upon Fgf15 overexpression. Plasma bile acid levels poorly correlate with bile acid pool, or hepatic bile acid uptake (18), but may largely be determined by intestinal transit (19), a process modulated by FGF19-signaling, independent of its proliferative effect (20). FGF15 induces genes involved in cell proliferation, including Ccnd1, Pcna and Cdkn1a. Restoring bile acid levels by cholate feeding (further) induced expression of these genes, but this effect was accompanied by even higher Fgf15 levels in both liver and intestine. Also, in conditions of partial hepatectomy genes reflecting proliferation were induced upon Fgf15 overexpression, both by overexpression in liver and intestine and by intestine-restricted overexpression. Incorporation of BrdU, which is a measure for proliferation, was only significantly increased in mice with both intestine and liver Fgf15 overexpression and not in the intestine-specific overexpression model. However, both overexpression models induced protein expression of CYCLIN D1, CDK4 and c-MYC. These analyzes suggest that FGF15 indeed activates cell cycle genes. Moreover, FGF15 knockout mice show a 70% mortality rate after partial hepatectomy (9), while AAV-mediated liver-specific Fgf15 overexpression in these mice could rescue this effect down to only 20% mortality. This implies that Fgf15 overexpression in liver also induced proliferative effects. Also, injections with recombinant FGF15 increased p-JNK, p-ERK and p-STAT3. Together, the data from Kong et al. suggests that FGF15 stimulates proliferation after hepatectomy independent of its effect on the bile acid pool size and largely independent of its production site. The latter is not entirely surprising since FGF15/19 have low heparin binding affinity, which allows them to diffuse far away from the intestine or liver and enables them to function as endocrine hormones (21).
The authors further investigated the mechanisms of action and studied effects of FGF15 on hepatocyte priming for regeneration, as well as termination of regeneration upon partial hepatectomy. Immediate-early response genes of c-Jun and c-Myc are significantly elevated after 30 and 60 minutes respectively in Fgf15 transgenic mice compared to wildtypes and this tendency was already present under basal (pre-hepatectomy) conditions. Furthermore, Fgf15 transgenic mice showed higher levels of phosphorylated Stat3, Nf-κΒ and IκΒα compared to wild-type mice. In addition, liver regeneration in wild-type mice was ongoing 7 days post-hepatectomy, and returned to their normal weight at day 14, whereas liver regeneration in mice overexpressing Fgf15 stopped at day 7, and the livers of Fgf15 overexpression mice were also smaller prior to the hepatectomy. Together, this indicates that FGF15 plays a role in priming during liver regeneration, but also in the termination process.
FGF15 and FGF19 only share a 51% amino acid sequence similarity (22). Peculiarly, the sequence divergence of FGF19 orthologues seems to be limited primarily to rodent FGF15 (23). Therefore, it seems important to study FGF15 in mice and FGF19 in humans, but to be careful with conclusions based on the injection or overexpression of FGF19 in mice. For instance, unlike FGF19, FGF15 does not cause accelerated tumor growth of hepatocellular carcinoma (HCC) in db/db mice or mice administered a high fat diet (24). Similarly, mice with humanized livers have a much larger livers (13% of body weight), compared with control mice, which was restored when the “humanized liver” mice also express FGF19, suggesting that hormone and its receptor need to be species-matched for their normal signaling (25). Therefore, new animal models like described in this study prove useful to further clarify the potential of FGF15/19 in liver regeneration.
Acknowledgements
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, AME Medical Journal. The article did not undergo external peer review.
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2018.09.08). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Michalopoulos GK. Liver regeneration. J Cell Physiol 2007;213:286-300. [Crossref] [PubMed]
- Marsh JW, Gray E, Ness R, et al. Complications of right lobe living donor liver transplantation. J Hepatol 2009;51:715-24. [Crossref] [PubMed]
- Taub R. Hepatoprotection via the IL-6/Stat3 pathway. J Clin Invest 2003;112:978-80. [Crossref] [PubMed]
- Inagaki T, Choi M, Moschetta A, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2005;2:217-25. [Crossref] [PubMed]
- Kir S, Beddow SA, Samuel VT, et al. FGF19 as a Postprandial, Insulin-Independent Activator of Hepatic Protein and Glycogen Synthesis. Science 2011;331:1621-4. [Crossref] [PubMed]
- Wu AL, Coulter S, Liddle C, et al. FGF19 Regulates Cell Proliferation, Glucose and Bile Acid Metabolism via FGFR4-Dependent and Independent Pathways. Plos One 2011;6. [PubMed]
- Miyake A, Nakayama Y, Konishi M, et al. Fgf19 regulated by Hh signaling is required for zebrafish forebrain development. Dev Biol 2005;288:259-75. [Crossref] [PubMed]
- Lin BC, Wang M, Blackmore C, et al. Liver-specific activities of FGF19 require Klotho beta. J Biol Chem 2007;282:27277-84. [Crossref] [PubMed]
- Kong B, Huang JS, Zhu Y, et al. Fibroblast growth factor 15 deficiency impairs liver regeneration in mice. Am J Physiol Gastrointest Liver Physiol 2014;306:G893-902. [Crossref] [PubMed]
- Pai R, Dunlap D, Qing J, et al. Inhibition of fibroblast growth factor 19 reduces tumor growth by modulating beta-catenin signaling. Cancer Res 2008;68:5086-95. [Crossref] [PubMed]
- Desnoyers L, Pai R, Ferrando R, et al. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene 2008;27:85-97. [Crossref] [PubMed]
- French DM, Lin BC, Wang MP, et al. Targeting FGFR4 Inhibits Hepatocellular Carcinoma in Preclinical Mouse Models. Plos One 2012;7. [PubMed]
- Zhou M, Wang XY, Phung V, et al. Separating Tumorigenicity from Bile Acid Regulatory Activity for Endocrine Hormone FGF19. Cancer Res 2014;74:3306-16. [Crossref] [PubMed]
- Inagaki T, Choi M, Moschetta A, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2005;2:217-25. [Crossref] [PubMed]
- Slijepcevic D, Roscam Abbing RL, Katafuchi T, et al. Hepatic uptake of conjugated bile acids is mediated by both sodium taurocholate cotransporting polypeptide and organic anion transporting polypeptides and modulated by intestinal sensing of plasma bile acid levels in mice. Hepatology 2017;66:1631-43. [Crossref] [PubMed]
- Kong B, Sun R, Huang M, et al. A Novel Fibroblast Growth Factor 15 Dependent- and Bile Acid Independent-Promotion of Liver Regeneration in Mice. Hepatology 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Schaap FG, van der Gaag NA, Gouma DJ, et al. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology 2009;49:1228-35. [Crossref] [PubMed]
- Slijepcevic D, Kaufman C, Wichers CG, et al. Impaired uptake of conjugated bile acids and hepatitis b virus pres1-binding in na(+) -taurocholate cotransporting polypeptide knockout mice. Hepatology 2015;62:207-19. [Crossref] [PubMed]
- Sips FL, Eggink HM, Hilbers PA, et al. In Silico Analysis Identifies Intestinal Transit as a Key Determinant of Systemic Bile Acid Metabolism. Front Physiol 2018;9:631. [Crossref] [PubMed]
- Oduyebo I, Camilleri M, Nelson AD, et al. Effects of NGM282, an FGF19 variant, on colonic transit and bowel function in functional constipation: a randomized phase 2 trial. Am J Gastroenterol 2018;113:725-34. [Crossref] [PubMed]
- Jones SA. Physiology of Fgf15/19. Adv Exp Med Biol 2012;728:171-82. [Crossref] [PubMed]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol 2001;2. [PubMed]
- Katoh M, Katoh M. FGF signaling network in the gastrointestinal tract Int J Oncol 2006;29:163-8. (Review). [PubMed]
- Zhou M, Luo J, Chen M, et al. Mouse species-specific control of hepatocarcinogenesis and metabolism by FGF19/FGF15. J Hepatol 2017;66:1182-92. [Crossref] [PubMed]
- Naugler WE, Tarlow BD, Fedorov LM, et al. Fibroblast Growth Factor Signaling Controls Liver Size in Mice With Humanized Livers. Gastroenterology 2015;149:728-40.e15. [Crossref] [PubMed]
Cite this article as: van de Wiel SMW, van de Graaf SFJ. Bile acid dependent and independent roles of fibroblast growth factor 15/19 in liver regeneration. AME Med J 2018;3:95.

