
The bile acid phospholipid conjugate ursodeoxycholate lysophoshatidylethanolamide acts by binding to calcium independent membrane phospholipase A2 type beta
Introduction
The actual challenge in hepatology is the therapy of non-alcoholic steatohepatitis (NASH). It presents as steatosis hepatitis with features of inflammation. NASH is the most common liver disease in the world. It develops to cirrhosis with its complications, including hepatocellular carcinoma. Several indicative laboratory parameters such as γ-glutamyl-transferase (GGT) and alanine aminotransferase (ALT) are elevated in this disease. However, a therapy is not available. Predisposing conditions are metabolic syndrome, diabetes and hypercholesterolemia. However, the pathogenesis is still unclear, but a key feature is the increased ratio of lysophosphatidylcholine (LPC) to phosphatidylcholine (PC), suggesting a higher activity of phospholipase A2 (PLA2) (1). The PLA2 family of phospholipases is classified into five distinct groups, namely the secretory (sPLA2), cytosolic (cPLA2), Ca2+-independent (iPLA2), lysosomal (LPLA2), and platelet-activating factor (PAF) acetylhydrolases (2). This complex family of enzymes plays a significant role in inflammation by triggering the synthesis of regulatory lipid molecules, which are produced and activated at the site of inflammation. Each member of this hydrolase enzyme family serves a distinct role by generating active lipid metabolites that promote inflammatory diseases including hyperlipidemia, obesity, and diabetes. In particular, the cPLA2s composed of different isoforms (PLA2α, PLA2β, and PLA2γ) participate in inflammation by hydrolyzing the ester bond at the sn-2 position of phospholipids to produce non-esterified fatty acids and lysophospholipids. In contrast, most sPLA2s contain a conserved calcium-binding loop and strongly dependent on Ca2+ concentrations for their catalytic activity (2). Members of the iPLA2 family have significant functions in transmembrane signal transduction, while the lysosomal PLA2 (known as PLA2G15) are involved in the catabolism of pulmonary surfactant and play a role in host defense and in the processing of lipid antigens for presentation by CD1 proteins (3). The three types of PAF acetylhydrolases, i.e., PAF-AH(I), PAF-AH(II), and PAF-AH(III) mediate the inactivation of the PAF by deacetylation of the acetyl group located at the sn-2 position of the glycerol backbone (4,5).
The PC breakdown product LPC is metabolically active for generation of phosphorylated C-Jun-N-terminal kinase 1 (pJNK1) (6). This key player accelerates cellular fatty acid influx (7) and induces inflammation and fibrosis. All downstream features of JNK1 signaling are well established (8). Nevertheless, the generation of LPC as precipitating event remains unresolved. Interestingly, hepatic fatty acid uptake is mediated by a heterotetrameric protein arrangement consisting of the fatty acid binding protein of plasma membranes (FABPPM) (9), the cluster of differentiation (CD36) (10), caveolin-1 (11), and calcium-independent membrane PLA2 type beta (iPLA2β) also known as group 6 iPLA2s (7). The complex is localized within detergent-resistant membrane platforms of the plasma membrane (DRM-PM) (7), representing specialized lipid domains constituting raft structures which mediate cellular communication (12). The iPLA2β is the prominent player in cellular PC hydrolysis (13). Within the fatty acid uptake complex, iPLA2β serves as the constitutive protein maintaining the required structural stability (14). It may not be directly involved in the fatty acid influx process, but it holds contact to intracellular metabolism for uptake regulation. It remains to be established, whether it may simply respond to cytosolic LPC concentration with feedback inhibition of its enzymatic activity or a more complex regulation takes place as previously proposed (15).
The bile acid phospholipid conjugate, ursodeoxycholate-lysophosphatidylethanolamide (UDCA-LPE) inhibits iPLA2β and suppresses pro-inflammatory LPC generation in a dose-dependent mode (7). According to the described dual function of iPLA2β as member of the fatty acid uptake complex and its intrinsic capability to generate LPC, UDCA-LPE is a potent inhibitor also of cellular fatty acid influx. Analysis of the uptake kinetics revealed a reduction of Vmax, which is compatible with fading of available active transport sites (7). Consequently, it reversed liver steatosis, inflammation, and fibrosis in animal studies, which are all critical features observed in NASH (16). Even TNF-α-induced acute liver failure could be prevented, if the drug was given concomitantly (17,18). Thus, UDCA-LPE could serve as hepatoprotective, anti-inflammatory drug for the liver (19). The open question remains, whether iPLA2β interacts with UDCA-LPE.
In the present study, we used in silico techniques for predicting potential docking sites of UDCA-LPE in iPLA2β. Our computational approach revealed a clear affinity of UDCA-LPE for iPLA2β with a region between Phe84 and Leu125 that should have the largest affinity for UDCA-LPE. Noteworthy, the proposed region is nearly identical with those that were determined for binding of pyrrophenone to the evolutionarily conserved PLA2α. Hence, the proposed direct binding of UDCA-LPE to iPLA2β might explain the efficacy of UDCA-LPE in preventing hepatic fatty acid uptake.
We present the following article in accordance with the handout on scientific writing and checklist of scientific manuscript components (available at https://amj.amegroups.com/article/view/10.21037/amj-21-10/rc).
Methods
General strategy used for molecular docking analysis
In order to analyze the binding energy and the docking pose of UDCA-LPE on iPLA2β protein from Chinese hamster (PDB ID: 6AUN), molecular docking was performed as described previously (20). Pyrrophenone (21) was used as positive control compound. The pyrrolidine derivative pyrrophenone is a cell-permeable highly potent and reversible inhibitor of the catalytic domain (CAT) cPLA2α through formation of a hemiketal between its ketone carbonyl and the active site serine of the enzyme based on the ability of polarized ketones to form stable hemiketals with serine proteases and esterases (22). For molecular docking analysis, this component is ideally suited, because its molecular mass is similar to UDCA-LPE. We used the following strategy for docking analysis. At first, unbiased blind docking was performed by covering the whole protein surface. The binding site of pyrrophenone was determined, and defined docking was performed by covering the binding site of pyrrophenone found after blind docking analysis. Then, we used the same setting for UDCA-LPE and identified potential interaction regions. In each case, the binding energy and docking pose of UDCA-LPE was compared with those of pyrrophenone.
Performance of docking analysis
For docking analysis, we used AutoDock 4.2, representing software with a graphical user interface offering a variety of search algorithms to explore a given docking problem (23). This automated docking algorithm is widely used for prediction of biomolecular complexes in structure/function analysis and in molecular design (23). It allows fully flexible modelling of specific portions of protein, in a similar manner as the ligand. In the algorithms, semi-empirical free energy force fields are used to predict the binding free energies of small molecules such as pyrrophenone or UDCA-LPE to supposed macromolecular target structures such as iPLA2β (23). The molecular graphics program Visual Molecular Dynamics (VMD), which is designed for the display and analysis of molecular assemblies (24), was used to visualize the docking poses.
Statistical analysis of docking prediction
In our analysis, the docking parameters were set to 250 runs and 2,500,000 energy evaluations for each cycle and three independent runs were performed by using AutoDock 4.2 algorithm (23). Average values of binding energies and standard deviations (SDs) were calculated and mentioned at the corresponding table.
Sequence alignments
Human, mouse, rat and Chinese hamster iPLA2β proteins were aligned using the Clustal Omega program (25). Sequence information from respective proteins were taken from the GenBank (Homo sapiens: NP_001336795.1; Mus musculus: NP_001185954; Rattus norvegicus: NP_001257725.1; Cricetulus griseus: 6AUN_B).
Remarks on scientific writing
We present the following article in accordance with the handout on scientific writing and checklist of scientific manuscript components according to the forms provided by the Teaching Issues and Experiments in Ecology (TIEE), which is a peer-reviewed web-based collection of ecological educational materials. The respective form can be found at: https://tiee.esa.org/vol/v4/experiments/habitat_shifts/pdf/Appendix3.pdf.
Results
iPLA2β, an evolutionarily conserved protein
The 85-kD cytosolic calcium-independent PLA2 (iPLA2β, also known as PLA2G6A or PNPLA9) was originally isolated from Chinese hamster ovary (CHO) cells (26). The full-length protein encodes an evolutionarily highly conserved 752-amino acid protein with one lipase motif (GXS465XG) and eight residue ankyrin repeats (Figure 1). It shares about 90.56% to 98.54% identity between human, Chinese hamster, mouse, and rat (Table 1).

Table 1
| Species | Human | Mouse | Rat | Hamster |
|---|---|---|---|---|
| Human | 100 | 91.09 | 90.96 | 90.56 |
| Mouse | 91.09 | 100 | 98.54 | 95.61 |
| Rat | 90.96 | 98.54 | 100 | 95.61 |
| Hamster | 90.56 | 95.61 | 95.61 | 100 |
iPLA2β, calcium-independent membrane phospholipase A2 type beta.
The enzyme belongs to a diverse group of enzymes catalyzing hydrolysis of the sn-2 substituent form glycerophospholipid substrates to yield a free fatty acid and a 2-lysophopholipid. There is now ample evidence that alterations in iPLA2β function is involved the pathogenesis of many diseases including cardiovascular disease, cancer, and muscular dystrophy (27). In addition, there is evidence that iPLA2β is involved in the pathogenesis of diabetes and non-alcoholic steatohepatitis (7,28). Therefore, iPLA2β inhibitors are potential good drug candidates to interfere with respective diseases. In line with this assumption, the selective inhibition of iPLA2β with a fluoroketone inhibitor (i.e., FKGK18) showed beneficial effects in models of autoimmune type 1 diabetes (29). We have recently shown that UDCA-LPE disintegrated the lipid backbone of raft plasma membrane domains by the removal of iPLA2β (14). In the mentioned study, we discussed that UDCA-LPE potentially exerted its effects by acting as an iPLA2β inhibitor. However, providing experimental proof for this hypothesis is complex, because interaction studies are mainly aggravated by the low solubility of UDCA-LPE and the fact that the CAT of the enzyme forms a tight dimer that is not easily accessible (27).
Prediction of interactions of iPLA2β UDCA-LPE
A well-established potent inhibitor for cPLA2α is pyrrophenone. This non-peptide and low-molecular weight inhibitor is structurally a 1,2,4-trisubstituted pyrrolidine that strongly inhibits the esterase and lysophospholipase activities of cellular PLA2α (30). Although structurally different, the overall size as measured by its molecular mass is similar to UDCA-LPE (Figure 2).
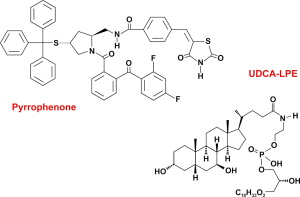
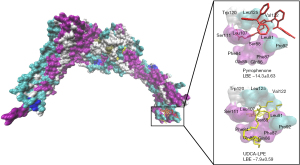
To get an impression of a possible binding of UDCA-LPE to iPLA2β, we performed a molecular docking approach using the pyrrophenone as a positive control compound. In an unbiased “blind docking” attempt, we found that potential interacting residues within iPLA2β for pyrrophenone are located between amino acid position Gln86 and Glu128. Upon performing the same setting for UDCA-LPE, we identified a region between Pro500 and Val678 that might have binding capacity. However, in a defined docking approach, in which potential pyrrophenone interacting residues were considered, we identified a region between Phe84 and Leu125 that should have the largest affinity for UDCA-LPE. The proposed region is nearly identical with those that were determined for pyrrophenone. The results for the blind and defined docking runs are summarized in Table 2.
Table 2
| Substance | LBE-1* | LBE-2* | LBE-3* | Mean | SD** | Interacting amino acid residues |
|---|---|---|---|---|---|---|
| Blind docking | ||||||
| UDCA-LPE | −1.45 | −0.34 | 0.35 | −0.48 | 0.91 | Pro500, Pro587, Thr588, Phe590, Arg591***, Pro592, Lys671, Lys675, Val678 |
| Pyrrophenone | −8.00 | −6.21 | −6.35 | −6.85 | 0.99 | Gln86, Ser88, Ser89, Leu91, Pro92, Leu107, Leu125, Ala126, Glu128 |
| Defined docking (pyrrophenone interacting residues in blind docking were considered) | ||||||
| UDCA-LPE | −8.38 | −7.24 | −8.09 | −7.90 | 0.59 | Phe84, Gln85, Gln86, Phe87, Ser88, Ser89, Leu91***, Pro92, Leu107, Gln108, Trp120, Thr121, Val122, Leu125 |
| Pyrrophenone | −13.99 | −15.07 | −13.97 | −14.34 | 0.63 | Phe84, Gln85, Phe87, Ser88***, Leu91, Pro92, Phe94, Val106, Leu107, Ser111***, Pro118, Ser119, Trp120***, Val122, Leu125, Ala126, Val127, Glu128 |
*, LBE: kcal/mol; **, average values of binding energies and standard deviations (SDs) are based on three independent AutoDock runs in which the docking parameters were set each to 250 runs and 2,500,000 energy evaluations; ***, residues that are making hydrogen bonds. UDCA-LPE, ursodeoxycholate-lysophosphatidylethanolamide; LBE, LBE, ligand-binding energy; PLA2, phospholipase A2.
It is remarkable that multiple sequence alignment for human, Chinese hamster, mouse, and rat iPLA2β protein revealed that the residues in the UDCA-LPE proposed docking site were all conserved for 10 out of 14 residues (cf. Figure 1) except for position His85 in humans, for position Tyr87 in humans and rats, for position Leu92 in humans, and for position Ser121 in humans. As shown in Figure 3, UDCA-LPE bound in close proximity to the pyrrophenone binding site but with lower affinity (−7.9±0.6 vs. −14.3±0.6 kcal/mol). Leu91 was forming a hydrogen bond with UDCA-LPE whereas Ser88, Ser111 and Trp120 were forming a hydrogen bond with pyrrophenone. Ten residues were commonly interacting with UDCA-LPE and pyrrophenone.
Discussion
The question addressed in this paper was, how UDCA-LPE exerts its inhibitory effect on fatty acid influx as well as inflammation of the liver. One option is based on its detergent effect on DRM-PM leading to dissolution of these platforms as prerequisite for constitution of the fatty acid uptake complex. In vitro experiments with isolated DRM-PM indeed showed that UDCA-LPE (50 µM) resulted in a 63.13%±7.40% reduction of phospholipids and an 81.94%±8.30% reduction of cholesterol in relation to mg total protein (14). The ratio of phospholipids to cholesterol changed from 2:1 to 4:1, resembling those of non-DRM fractions (31). Thus, iPLA2β could lose contact to DRM-PM and distribute to other cell compartments.
The other option is that iPLA2β is primarily and specifically removed from the membrane fatty acid uptake complex. This is followed by removal of the other members of the fatty acid uptake complex (7,14). Moreover, if PC remains bound to iPLA2β even if iPLA2β is displaced from DRM-PMs and the enzymatic activity is silenced after UDCA-LPE exposure, it is likely that the DRM-PM lipid platform is consequently dissolved (14).
In favor of the iPLA2β removal option is the observation that the same phenomenon was observed with other phospholipase inhibitors, which are not considered as detergents: methyl arachidonyl fluorophosphonate (MAFP) and bromoenol lactone (BEL) (7). Therefore, the next question, which had to be answered, is whether UDCA-LPE binds to iPLA2β. Recently the structure of iPLA2β was resolved, and it was suggested that this protein forms a stable dimer, in which the active sites of the dimer are wide open providing sufficient space for phospholipids to access the catalytic centers (27). Moreover, the enzymatic activity of the dimer can be allosterically inhibited by calmodulin altering the confirmation of the dimerization interface (27). The question arises, whether dimerization is obligatory for its enzymatic function and whether monomeric iPLA2β within the heterotetrameric fatty acid uptake complex serves for the same activation purpose. One could argue that the subunit of iPLA2β communicates via ankyrin repeats with other proteins (15). Indeed, mutations lacking the ankyrin repeats of iPLA2β retain their CAT, but are catalytically inactive (15).
In this study, we demonstrated that UDCA-LPE bound with high affinity to iPLA2β. Since the UDCA-LPE binding energy was lower than −7 kcal/mol, it reflects binding affinity. According to the literature, −7 kcal/mol can be considered as threshold value reflecting strong interaction with the protein of interest (32-34). Based on theoretical considerations and respective modeling systems, the applied molecular docking approach is a valuable tool to provide an estimate of the location and binding energy of a ligand toward a target protein. In contrast, a direct experimental evaluation of UDCA-LPE binding is prone to artifacts, because iPLA2β is embedded in DRM-PM platforms and the required purification disturbs its structure and, thus, its function.
Next, it has to be addressed, whether UDCA-LPE leads to allosteric inhibition of iPLA2β via conformational change, in a comparable manner as reported for MAFP and BEL (15). As shown in Figure 3, UDCA-LPE binding is proposed outside the catalytic side of the enzyme, which is compatible with a concomitant structural change. If the requirements for binding to the enzyme are defined, other small molecules can be designed for optimal clinical use. Bioavailability, effectiveness, metabolic disposition, adverse events and toxicity are the relevant evaluation criteria.
These findings are of fundamental importance and may have direct therapeutic implications. UDCA-LPE is a potent inhibitor of cellular fatty acid influx and a direct inhibitor of iPLA2β activity. As such, compounds with similar structure to UDCA-LPE but overall higher solubility might be an effective therapeutic mean for NASH therapy. It will be interesting to follow up, how the drug-likeness of these compounds could be translated into novel therapies.
Our study has some limitations. For docking analysis, we used a template consisting of pyrrophenone and cPLA2α for determination of binding sites of UDCA-LPE in iPLA2β. Although both proteins belong to the phospholipase superfamily, it is obvious that they have defined functions that might predict different structures. However, sophisticated structure analysis has shown that the fold of the core secondary structure elements in the CAT of iPLA2β resembles that of cPLA2α CAT (27), which suggests that cPLA2α is a good template protein. Likewise, the cPLA2α inhibitor pyrrophenone (CAS no. 341973-06-6) has a molecular weight of ~850 and molecular formula C49H37F2N3O5S2, while the composite of ursodeoxycholic acid (CAS no. 128-13-2; molecular weight ~393, C24H40O4) and lysophosphatidylethanolamine (CAS no. 95046-40-5; molecular weight ~479; C7H15NO7P−) have a different molecular structure and, thus, mobility that might interfere with the dynamics in binding to its predicted target. Moreover, the water solubility of pyrrophenone and UDCA-LPE may vary, which per se might influence the binding to their target sites. UDCA-LPE has limited solubility and the cell-permeable inhibitor pyrrophenone is only sparingly soluble in aqueous buffers too (35,36). This suggests that the solubility in our assay may affect both compound-enzyme interactions. Unfortunately, the low solubility of UDCA-LPE hindered us to document the binding of UDCA-LPE to iPLA2β, because UDCA-LPE formed micelles in water, which prevented effective testing of the proposed interaction by techniques such as surface plasmon resonance, microscale thermophoresis, or isothermal titration calorimetry.
In case an optional iPLA2β inhibitor is found, the perspective for development of an anti-inflammatory, antisteatotic and antifibrotic drug for NASH and other inflammatory liver diseases is opened.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, AME Medical Journal for the series “Liver Diseases: Symptoms, Causes and Novel Therapies”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the handout on scientific writing and checklist of scientific manuscript components according to the forms provided by the Teaching Issues and Experiments in Ecology (TIEE), which is a peer-reviewed web-based collection of ecological educational materials. Available at https://amj.amegroups.com/article/view/10.21037/amj-21-10/rc
Data Sharing Statement: Available at https://amj.amegroups.com/article/view/10.21037/amj-21-10/dss
Peer Review File: Available at https://amj.amegroups.com/article/view/10.21037/amj-21-10/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://amj.amegroups.com/article/view/10.21037/amj-21-10/coif). The series “Liver Diseases: Symptoms, Causes and Novel Therapies” was commissioned by the editorial office without any funding or sponsorship. WS and RW served as unpaid Guest Editors of the series. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Law SH, Chan ML, Marathe GK, et al. An updated review of lysophosphatidylcholine metabolism in human diseases. Int J Mol Sci 2019;20:1149. [Crossref] [PubMed]
- Filkin SY, Lipkin AV, Fedorov AN. Phospholipase superfamily: Structure, functions, and biotechnological applications. Biochemistry (Mosc) 2020;85:S177-95. [Crossref] [PubMed]
- Shayman JA, Tesmer JJG. Lysosomal phospholipase A2. Biochim Biophys Acta Mol Cell Biol Lipids 2019;1864:932-40. [Crossref] [PubMed]
- Arai H, Koizumi H, Aoki J, et al. Platelet-activating factor acetylhydrolase (PAF-AH). J Biochem 2002;131:635-40. [Crossref] [PubMed]
- Kono N, Arai H. Platelet-activating factor acetylhydrolases: An overview and update. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:922-31. [Crossref] [PubMed]
- Fang X, Gibson S, Flowers M, et al. Lysophosphatidylcholine stimulates activator protein 1 and the c-Jun N-terminal kinase activity. J Biol Chem 1997;272:13683-9. [Crossref] [PubMed]
- Stremmel W, Staffer S, Wannhoff A, et al. Plasma membrane phospholipase A2 controls hepatocellular fatty acid uptake and is responsive to pharmacological modulation: implications for nonalcoholic steatohepatitis. FASEB J 2014;28:3159-70. [Crossref] [PubMed]
- Seki E, Brenner DA, Karin M. A liver full of JNK: signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology 2012;143:307-20. [Crossref] [PubMed]
- Stremmel W, Strohmeyer G, Borchard F, et al. Isolation and partial characterization of a fatty acid binding protein in rat liver plasma membranes. Proc Natl Acad Sci U S A 1985;82:4-8. [Crossref] [PubMed]
- Abumrad NA, el-Maghrabi MR, Amri EZ, et al. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J Biol Chem 1993;268:17665-8. [Crossref] [PubMed]
- Trigatti BL, Anderson RG, Gerber GE. Identification of caveolin-1 as a fatty acid binding protein. Biochem Biophys Res Commun 1999;255:34-9. [Crossref] [PubMed]
- Simons K, Ehehalt R. Cholesterol, lipid rafts, and disease. J Clin Invest 2002;110:597-603. [Crossref] [PubMed]
- Hermansson M, Hänninen S, Hokynar K, et al. The PNPLA-family phospholipases involved in glycerophospholipid homeostasis of HeLa cells. Biochim Biophys Acta 2016;1861:1058-65. [Crossref] [PubMed]
- Stremmel W, Staffer S, Fricker G, et al. The bile acid-phospholipid conjugate ursodeoxycholyl-lysophosphatidylethanolamide (UDCA-LPE) disintegrates the lipid backbone of raft plasma membrane domains by the removal of the membrane phospholipase A2. Int J Mol Sci 2019;20:5631. [Crossref] [PubMed]
- Ramanadham S, Ali T, Ashley JW, et al. Calcium-independent phospholipases A2 and their roles in biological processes and diseases. J Lipid Res 2015;56:1643-68. [Crossref] [PubMed]
- Pathil A, Mueller J, Warth A, et al. Ursodeoxycholyl lysophosphatidylethanolamide improves steatosis and inflammation in murine models of nonalcoholic fatty liver disease. Hepatology 2012;55:1369-78. [Crossref] [PubMed]
- Pathil A, Warth A, Chamulitrat W, et al. The synthetic bile acid-phospholipid conjugate ursodeoxycholyl lysophosphatidylethanolamide suppresses TNFalpha-induced liver injury. J Hepatol 2011;54:674-84. [Crossref] [PubMed]
- Chamulitrat W, Zhang W, Xu W, et al. Hepatoprotectant ursodeoxycholyl lysophosphatidylethanolamide increasing phosphatidylcholine levels as a potential therapy of acute liver injury. Front Physiol 2012;3:24. [Crossref] [PubMed]
- Chamulitrat W, Burhenne J, Rehlen T, et al. Bile salt-phospholipid conjugate ursodeoxycholyl lysophosphatidylethanolamide as a hepatoprotective agent. Hepatology 2009;50:143-54. [Crossref] [PubMed]
- Kadioglu O, Saeed M, Kuete V, et al. Oridonin targets multiple drug-resistant tumor cells as determined by in silico and in vitro analyses. Front Pharmacol 2018;9:355. [Crossref] [PubMed]
- Kurosawa T, Nakamura H, Yamaura E, et al. Cytotoxicity induced by inhibition of thioredoxin reductases via multiple signaling pathways: role of cytosolic phospholipase A(2)alpha-dependent and -independent release of arachidonic acid. J Cell Physiol 2009;219:606-16. [Crossref] [PubMed]
- Yun B, Lee H, Ewing H, et al. Off-target effect of the cPLA2α inhibitor pyrrophenone: Inhibition of calcium release from the endoplasmic reticulum. Biochem Biophys Res Commun 2016;479:61-6. [Crossref] [PubMed]
- Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785-91. [Crossref] [PubMed]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996;14:33-8, 27-8.
- Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539. [Crossref] [PubMed]
- Tang J, Kriz RW, Wolfman N, et al. A novel cytosolic calcium-independent phospholipase A2 contains eight ankyrin motifs. J Biol Chem 1997;272:8567-75. [Crossref] [PubMed]
- Malley KR, Koroleva O, Miller I, et al. The structure of iPLA. Nat Commun 2018;9:765. [Crossref] [PubMed]
- Lei X, Zhang S, Emani B, et al. A link between endoplasmic reticulum stress-induced β-cell apoptosis and the group VIA Ca2+-independent phospholipase A2 (iPLA2β). Diabetes Obes Metab 2010;12:93-8. [Crossref] [PubMed]
- Bone RN, Gai Y, Magrioti V, et al. Inhibition of Ca2+-independent phospholipase A2β (iPLA2β) ameliorates islet infiltration and incidence of diabetes in NOD mice. Diabetes 2015;64:541-54. [Crossref] [PubMed]
- Ono T, Yamada K, Chikazawa Y, et al. Characterization of a novel inhibitor of cytosolic phospholipase A2alpha, pyrrophenone. Biochem J 2002;363:727-35. [Crossref] [PubMed]
- Zidovetzki R, Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim Biophys Acta 2007;1768:1311-24. [Crossref] [PubMed]
- Kadioglu O, Efferth T. Peptide aptamer identified by molecular docking targeting translationally controlled tumor protein in leukemia cells. Invest New Drugs 2016;34:515-21. [Crossref] [PubMed]
- Aubin-Tam ME, Appleyard DC, Ferrari E, et al. Adhesion through single peptide aptamers. J Phys Chem A 2011;115:3657-64. [Crossref] [PubMed]
- Chushak Y, Stone MO. In silico selection of RNA aptamers. Nucleic Acids Res 2009;37:e87. [Crossref] [PubMed]
- Maharjan P, Kim D, Jin M, et al. Preclinical evaluation of UDCA-containing oral formulation in mice for the treatment of wet age-related macular degeneration. Pharmaceutics 2019;11:561. [Crossref] [PubMed]
- Avadhani M, Geyer R, White DC, et al. Lysophosphatidylethanolamine is a substrate for the short-chain alcohol dehydrogenase SocA from Myxococcus xanthus. J Bacteriol 2006;188:8543-50. [Crossref] [PubMed]
Cite this article as: Stremmel C, Stremmel W, Kadioglu O, Efferth T, Weiskirchen R. The bile acid phospholipid conjugate ursodeoxycholate lysophoshatidylethanolamide acts by binding to calcium independent membrane phospholipase A2 type beta. AME Med J 2021;6:24.

