
Cancer detection and tissue of origin determination with novel annotation and scoring of cell-free methylated DNA
In a recent paper, Guo et al. (1) analysed the methylation of DNA fragments circulating in the blood plasma that are released when cells die. Their analysis used a novel approach to annotate DNA methylation and a methylation status score, which captures both average methylation level and the pattern of co-methylation. They were able to detect lung and colorectal cancers and to predict their tissue of origin.
Blood plasma contains cell-free DNA (cfDNA) originating from dead cells. This cfDNA is present at low concentrations (2) and increases when there is necrotic tissue in which more cells than usual are lysing and releasing their DNA (3). This cfDNA is fragmented into pieces with a modal length of 166 bp, approximately the size of DNA that is associated with a single nucleosomal unit (4). cfDNA in blood plasma has the potential to be a biomarker with strong clinical utility for diseases, such as cancer, as it is derived from almost all of the tissues of the body and is easily accessible through venepuncture. Therefore, cfDNA may be used to assess cancer in tissues that are not easily accessible for biopsy and monitor cancer progression, remission, and relapse over multiple time points. Abbosh et al. (5) have recently explored the application of cfDNA to track the evolutionary dynamics of early-stage tumours.
Epigenetic control of gene expression includes the methylation of DNA, especially in CpG dinucleotides. Specific patterns of DNA methylation are frequently characteristic of a given tissue. Consequently, analysing the DNA methylation pattern of a length of DNA can be used to predict from which tissue it originated (6). In addition to the mutational changes affecting the cancer genome, the DNA methylation patterns of cancer cells are highly disrupted, undergoing a global loss of DNA methylation as well as hypermethylation in specific regions including some tumour suppressor genes (7). Changes in the underlying genetic sequence can also produce alterations in the DNA methylome. Thus, Guo et al. (1) have examined the methylation of cfDNA in an attempt to detect cancer and predict its tissue of origin (Figure 1).
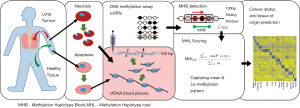
The methylation status of CpG sites adjacent to one another in the genome is correlated (8) and is strongly predicted by CpG density (9). Groups of CpGs are frequently more informative about the functional status of their genomic locus than are individual CpG sites (10). In order to annotate the cfDNA methylation profiles with features that would provide more useful information, the authors drew an analogy between these correlated methylation states and the correlation of genotypes at adjacent loci. They made use of the same mathematics underpinning the concept of linkage disequilibrium (LD) in order to annotate co-methylation blocks, which they termed methylation haplotype blocks (MHBs). In order to score these features according to both their methylation level and their degree of co-methylation, they described a metric called methylation haplotype load (MHL). MHL is a weighted mean of the fraction of all possible fully methylated substrings of the region for which the MHL is being measured. Importantly MHL is able to distinguish between features with the same methylation levels and different degrees of co-methylation.
Using 61 whole-genome bisulfite sequencing (WGBS) datasets drawn from human primary tissues, primary tumour samples, progenitor cells and cancer cell lines the authors identified 147,888 MHBs. These MHBs had an average size of 95 bp, and contained a minimum of 3 CpG sites per block, representing ~0.5% of the human genome. Most MHBs exhibit perfect coupling of their constituent CpGs, with r2>0.9 in 94.8% of MHBs in cultured stem and progenitor cells; 91.2% of MHBs in somatic cells from mixed primary adult tissue; and 87.8% of MHBs from mixed colorectal and lung cancer tissues and cell lines. This decline in the correlation of CpG from stem/progenitor cells, through somatic tissue, to cancer cells is consistent with previous observations of increasing disorder in methylation across these tissues (11). 41.1% of MHBs were intergenic while 58.9% are within transcribed regions. MHBs were enriched in functional loci, including enhancers, promoters, CpG islands and variably methylated regions. To calibrate their cancer status prediction models, they looked at the MHL of MHBs in 158 reduced representation bisulfite sequencing (RRBS) data from healthy and disease, plasma and primary tissue datasets. They parameterised their tissue of origin prediction model with 43 mixed WGBS and RRBS samples from 10 healthy human tissues.
This cfDNA methylation study exploited techniques for the epigenetic analysis of small quantities of DNA developed for single-cell sequencing (scRRBS) (12). The authors were able to detect lung and colorectal cancer and identify their tissue of origin. They identified colorectal cancer and lung cancer with 96.7% and 93.1% sensitivity, and 94.6% and 90.6% specificity, respectively. In the detection of cancer, the MHL metric outperformed mean methylation and individual CpG methylation scores of MHBs. Samples were classified according to their tissue of origin with an accuracy for colorectal cancer of 82.8%, lung cancer of 88.5% and healthy tissue of 91.2%. Tissue of origin classification performance was best when limited to tumour samples of less heterogeneous clinical status.
The analysis of small quantities of DNA present in cfDNA shares many of the same limitations as does single cell DNA methylation profiling. Specifically, there is a tendency to get reasonable quality data for a quasi-random subset of the genome with other areas left essentially devoid of useful information. Therefore, there may not be data on the regions that possess the most predictive value in the models.
Whilst the authors found support for their MHBs through higher correlation of CpGs within MHBs in RRBS and even Illumina Infinium HumanMethylation450 BeadChip (450 k array) data, their initial WGBS discovery dataset was quite small given the number of tissues that they examined. Additional data will be needed to establish which MHBs are the most robust and reproducible. The MHB features and MHL metric developed here may have potentially useful applications in the wider study of DNA methylation. MHBs may be able to identify functional units of methylation in a way that differs from existing approaches such as those based on CpG density, differentially methylated regions and differentially variable regions. MHL scores could be used in place of average methylation levels of features in contexts where both methylation level and the extent co-methylation are pertinent.
At this point, the sensitivity and specificity of this method as a diagnostic test for cancer and the accuracy of tissue of origin prediction are too low for application in most clinical settings. However, this model was constructed on a relatively sparse dataset and more extensive training data may produce improvements in predictive quality. As the authors noted, an additional area to investigate will be to attempt training of a model on blood samples from patients drawn at various time points prior to cancer diagnosis to assess the utility of this method in early detection. Given the level of accuracy that was achieved with the available data, along with the ever-declining costs and increasing performance of the technologies involved, this approach could become practicable for use in clinical settings in the relatively near future.
Acknowledgements
Funding: RJ Acton and CG Bell acknowledge funding support from the MRC (U.K.) via the MRC Lifecourse Epidemiology Unit.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. An-Qiang Wang (Department of Liver Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China).
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/amj.2017.08.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Guo S, Diep D, Plongthongkum N, et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat Genet 2017;49:635-42. [Crossref] [PubMed]
- Page K, Guttery DS, Zahra N, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 2013;8:e77963. [Crossref] [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed]
- Ulz P, Thallinger GG, Auer M, et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat Genet 2016;48:1273-8. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Moran S, Martínez-Cardús A, Sayols S, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol 2016;17:1386-95. [Crossref] [PubMed]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 2007;8:286-98. [Crossref] [PubMed]
- Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 2006;38:1378-85. [Crossref] [PubMed]
- Baubec T, Schübeler D. Genomic patterns and context specific interpretation of DNA methylation. Curr Opin Genet Dev 2014;25:85-92. [Crossref] [PubMed]
- Ziller MJ, Gu H, Müller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013;500:477-81. [Crossref] [PubMed]
- Jenkinson G, Pujadas E, Goutsias J, et al. Potential energy landscapes identify the information-theoretic nature of the epigenome. Nat Genet 2017;49:719-29. [Crossref] [PubMed]
- Worm Ørntoft MB, Jensen SØ, Hansen TB, et al. Comparative analysis of 12 different kits for bisulfite conversion of circulating cell-free DNA. Epigenetics 2017;1-11. [Crossref] [PubMed]
Cite this article as: Acton RJ, Bell CG. Cancer detection and tissue of origin determination with novel annotation and scoring of cell-free methylated DNA. AME Med J 2017;2:110.

